2: Structuring¶
Here you'll find more info about creating and using beep to do your own custom cycler analyses.
BEEPDatapath- One object for ingestion, structuring, and validation- Batch functions for structuring
- Featurization
- Running and analyzing models
Structuring with BEEPDatapath¶
One class for ingestion, structuring, and validation¶
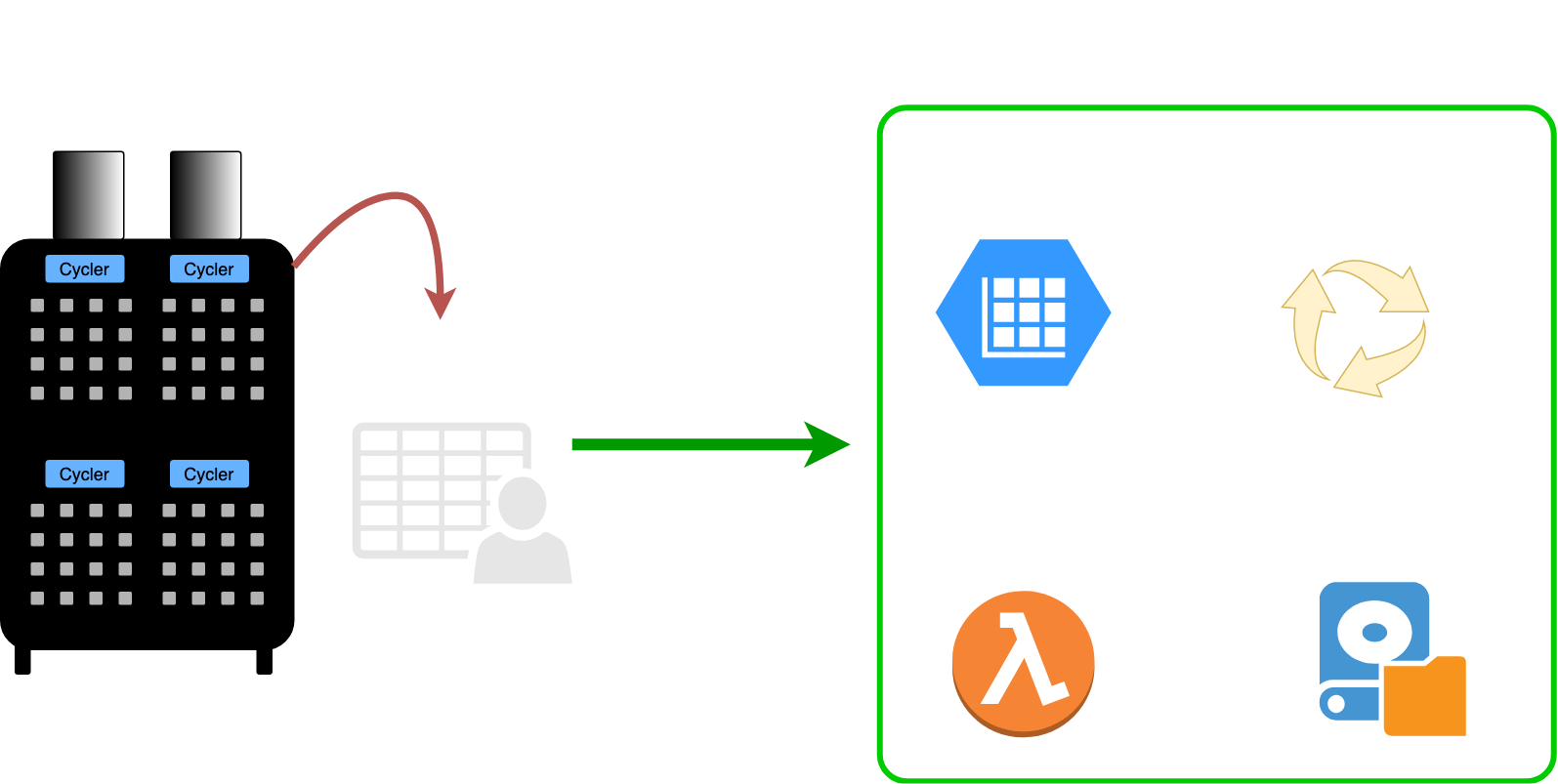
BEEPDatapath is an abstract base class that can handle ingestion, structuring, and validation for many types of cyclers. A datapath
object represents a complete processing pipeline for battery cycler data.
Each cycler has it's own BEEPDatapath class:
ArbinDatapathMaccorDatapathNewareDatapathIndigoDatapathBiologicDatapath
All these datapaths implement the same core methods, properties, and attributes, listed below:
Methods for loading and serializing battery cycler data¶
*Datapath.from_file(filename)¶
Classmethod to load a raw cycler output file (e.g., a csv) into a datapath object. Once loaded, you can validate or structure the file.
# Here we use ArbinDatapath as an example
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("my_arbin_file.csv")
*Datapath.to_json_file(filename)¶
Dump the current state of a datapath to a file. Can be later loaded with from_json_file.
from beep.structure import NewareDatapath
datapath = NewareDatapath.from_file("/path/to/my_raw_neware_file")
# do some operations
...
# Write the processed file to disk, which can then be loaded.
datapath.to_json_file("my_processed_neware_data.json")
*Datapath.from_json_file(filename)¶
Classmethod to load a processed cycler file (e.g., a previously structured Datapath) into a datapath object.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_json_file("my_previously_serialized_datapath.json")
*Datapath(data, metadata, paths=None, **kwargs)¶
Initialize any cycler from the raw data (given as a pandas dataframe) and metadata (given as a dictionary). Paths can be included to keep track of where various cycler files are located. Note: This is not the recommended way to create a BEEPDatapath, as data and metadata must have specific formats to load and structure correctly.
Validation and structuring with BEEPDatapaths¶
*Datapath.validate()¶
Validate your raw data. Will return true if the raw data is valid for your cycler (i.e., can be structured successfully).
from beep.structure import IndigoDatapath
datapath = IndigoDatapath.from_file("/path/to/my_indigo_file")
is_valid = datapath.validate()
print(is_valid)
# Out:
# True or False
*Datapath.structure(*args)¶
Interpolate and structure your data using specified arguments. Once structured, your BEEPDatapath is able to access things like the diagnostic summary, interpolated cycles, cycle summary, diagnostic summary, cycle life, and more (see Analysis and attributes of core attributes of BEEPDatapath)
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("my_arbin_file.csv")
# Structure your data by manually specifying parameters.
datapath.structure(v_range=[1.2, 3.5], nominal_capacity=1.2, full_fast_charge=0.85)
*Datapath.autostructure()¶
Run structuring using automatically determined parameters. BEEP can automatically detect the structuring parameters based on your raw data.
Note: The BEEP environment variable BEEP_PROCESSING_DIR must be set before autostructuring, and this directory must contain a parameters file which can be used for determine_structuring_parameters.
from beep.structure import BiologicDatapath
datapath = BiologicDatapath.from_file("path/to/my/biologic_data_file")
# Automatically determines structuring parameters and structures data
datapath.autostructure()
Analysis and core attributes of BEEPDatapath¶
*Datapath.paths¶
Access all paths of files related to this datapath. paths is a simple mapping of {file_description: file_path} which holds the paths of all files related to this datapath, including raw data, metadata, EIS files, and structured outputs.
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("/path/to/my_arbin_file.csv")
print(datapath.paths)
# Out:
{"raw": "/path/to/my_arbin_file.csv", "metadata": "/path/to/my_arbin_file_Metadata.csv"}
*Datapath.structuring_parameters¶
Parameters used to structure BEEPDatapaths:
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("/path/to/my_arbin_file.csv")
datapath.autostructure()
print(datapath.structuring_parameters)
# Out:
{'v_range': None,
'resolution': 1000,
'diagnostic_resolution': 500,
'nominal_capacity': 1.1,
'full_fast_charge': 0.8,
'diagnostic_available': False,
'charge_axis': 'charge_capacity',
'discharge_axis': 'voltage'}
*Datapath.raw_data¶
The raw data, loaded into a standardized dataframe format, of this datapath's battery cycler data.
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("/path/to/my_arbin_file.csv")
print(datapath.raw_data)
# Out:
data_point test_time ... temperature date_time_iso
0 0 0.0021 ... 20.750711 2017-12-05T03:37:36+00:00
1 1 1.0014 ... 20.750711 2017-12-05T03:37:36+00:00
2 2 1.1165 ... 20.750711 2017-12-05T03:37:36+00:00
3 3 2.1174 ... 20.750711 2017-12-05T03:37:36+00:00
4 4 12.1782 ... 20.750711 2017-12-05T03:37:36+00:00
... ... ... ... ... ...
251258 251258 30545.2000 ... 32.595604 2017-12-14T00:10:40+00:00
251259 251259 30545.2000 ... 32.555054 2017-12-14T00:10:40+00:00
251260 251260 30550.1970 ... 32.555054 2017-12-14T00:12:48+00:00
251261 251261 30550.1970 ... 32.545870 2017-12-14T00:12:48+00:00
251262 251262 30555.1970 ... 32.445827 2017-12-14T00:12:48+00:00
*Datapath.metadata¶
An object holding all metadata for this datapath's cycler run.
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("/path/to/my_arbin_file.csv")
print(datapath.metadata.barcode)
print(datapath.metadata.channel_id)
print(datapath.metadata.protocol)
print(datapath.metadata.raw)
# Out:
"EL151000429559"
28
'2017-12-04_tests\\20170630-4_65C_69per_6C.sdu'
{'test_id': 296, 'device_id': 60369369, 'channel_id': 28, 'start_datetime': 1512445026, '_resumed_times': 0, 'last_resume_datetime': 0, '_last_end_datetime': 1512514129, 'protocol': '2017-12-04_tests\\20170630-4_65C_69per_6C.sdu', '_databases': 'ArbinResult_43,ArbinResult_44,ArbinResult_45,', 'barcode': 'EL151000429559', '_grade_id': 0, '_has_aux': 3, '_has_special': 0, '_schedule_version': 'Schedule Version 7.00.08', '_log_aux_data_flag': 1, '_log_special_data_flag': 0, '_rowstate': 0, '_canconfig_filename': nan, '_m_ncanconfigmd5': nan, '_value': 0.0, '_value2': 0.0}
*Datapath.structured_data¶
The structured (interpolated) data, as a dataframe. The format is similar to that of .raw_data. The datapath must be structured before this attribute is available.
from beep.structure import ArbinDatapath
datapath = ArbinDatapath.from_file("/path/to/my_arbin_file.csv")
datapath.autostructure()
print(datapath.structured_data)
# Out:
voltage test_time current ... temperature cycle_index step_type
0 2.500000 NaN NaN ... NaN 0 discharge
1 2.501702 NaN NaN ... NaN 0 discharge
2 2.503403 NaN NaN ... NaN 0 discharge
3 2.505105 NaN NaN ... NaN 0 discharge
4 2.506807 NaN NaN ... NaN 0 discharge
... ... ... ... ... ... ...
461995 NaN NaN NaN ... NaN 245 charge
461996 NaN NaN NaN ... NaN 245 charge
461997 NaN NaN NaN ... NaN 245 charge
461998 NaN NaN NaN ... NaN 245 charge
461999 NaN NaN NaN ... NaN 245 charge
*Datapath.structured_summary¶
A summary of the structured cycler data, as a dataframe. The datapath must be structured before this attribute is available.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file.071")
datapath.autostructure()
print(datapath.structured_summary)
# Out:
cycle_index discharge_capacity charge_capacity discharge_energy charge_energy dc_internal_resistance temperature_maximum temperature_average temperature_minimum date_time_iso energy_efficiency charge_throughput energy_throughput charge_duration time_temperature_integrated paused
cycle_index
0 0 4.719281 3.827053 17.273731 14.901985 0.0 NaN NaN NaN 2019-12-17T17:51:51+00:00 1.159156 3.827053 14.901985 NaN NaN 4957
6 6 2.074518 4.406801 7.677041 16.997186 0.0 NaN NaN NaN 2019-12-20T13:14:40+00:00 0.451665 8.233854 31.899172 5791.0 NaN 0
7 7 2.097911 2.108322 7.775166 8.597635 0.0 NaN NaN NaN 2019-12-20T15:51:34+00:00 0.904338 10.342176 40.496807 NaN NaN 0
8 8 2.074545 2.098428 7.684986 8.557546 0.0 NaN NaN NaN 2019-12-20T17:32:21+00:00 0.898036 12.440605 49.054352 NaN NaN 0
9 9 2.074061 2.082069 7.685348 8.494265 0.0 NaN NaN NaN 2019-12-20T19:12:46+00:00 0.904769 14.522674 57.548618 NaN NaN 0
10 10 2.065671 2.069061 7.655246 8.441246 0.0 NaN NaN NaN 2019-12-20T20:52:53+00:00 0.906886 16.591734 65.989861 NaN NaN 0
11 11 2.064542 2.068921 7.651949 8.439011 0.0 NaN NaN NaN 2019-12-20T22:32:38+00:00 0.906735 18.660656 74.428871 NaN NaN 0
12 12 2.068333 2.061454 7.666199 8.409441 0.0 NaN NaN NaN 2019-12-21T00:12:35+00:00 0.911618 20.722109 82.838318 NaN NaN 0
13 13 2.054566 2.067370 7.616584 8.431127 0.0 NaN NaN NaN 2019-12-21T01:52:14+00:00 0.903389 22.789478 91.269440 NaN NaN 0
14 14 2.061369 2.057715 7.647454 8.394535 0.0 NaN NaN NaN 2019-12-21T03:31:54+00:00 0.911004 24.847195 99.663979 NaN NaN 0
15 15 2.050721 2.059819 7.602874 8.401562 0.0 NaN NaN NaN 2019-12-21T05:11:24+00:00 0.904936 26.907013 108.065536 NaN NaN 0
16 16 2.055427 2.057405 7.622452 8.393292 0.0 NaN NaN NaN 2019-12-21T06:50:57+00:00 0.908160 28.964418 116.458832 NaN NaN 0
17 17 2.045344 2.049606 7.583858 8.360918 0.0 NaN NaN NaN 2019-12-21T08:30:36+00:00 0.907060 31.014025 124.819748 NaN NaN 0
18 18 2.047280 2.046608 7.591624 8.347446 0.0 NaN NaN NaN 2019-12-21T10:09:56+00:00 0.909455 33.060631 133.167191 NaN NaN 0
19 19 2.055454 2.046478 7.623849 8.347916 0.0 NaN NaN NaN 2019-12-21T11:49:18+00:00 0.913264 35.107109 141.515106 NaN NaN 0
20 20 2.043676 2.055780 7.579766 8.383341 0.0 NaN NaN NaN 2019-12-21T13:28:39+00:00 0.904146 37.162891 149.898453 NaN NaN 0
21 21 2.049323 2.046085 7.605977 8.346517 0.0 NaN NaN NaN 2019-12-21T15:08:10+00:00 0.911276 39.208977 158.244965 NaN NaN 0
22 22 2.038514 2.047097 7.560916 8.349430 0.0 NaN NaN NaN 2019-12-21T16:47:22+00:00 0.905561 41.256073 166.594406 NaN NaN 0
23 23 2.044779 2.045038 7.585164 8.342201 0.0 NaN NaN NaN 2019-12-21T18:26:38+00:00 0.909252 43.301109 174.936600 NaN NaN 0
24 24 2.039805 2.039563 7.567169 8.319416 0.0 NaN NaN NaN 2019-12-21T20:06:10+00:00 0.909579 45.340672 183.256012 NaN NaN 0
25 25 2.039563 2.040318 7.566332 8.320876 0.0 NaN NaN NaN 2019-12-21T21:45:20+00:00 0.909319 47.380993 191.576889 NaN NaN 0
26 26 2.052362 2.038989 7.616830 8.316606 0.0 NaN NaN NaN 2019-12-21T23:24:33+00:00 0.915858 49.419979 199.893494 NaN NaN 0
27 27 2.035744 2.051446 7.552814 8.364671 0.0 NaN NaN NaN 2019-12-22T01:03:48+00:00 0.902942 51.471428 208.258163 NaN NaN 0
28 28 2.039347 2.041048 7.568011 8.325755 0.0 NaN NaN NaN 2019-12-22T02:43:14+00:00 0.908988 53.512474 216.583923 NaN NaN 0
*Datapath.diagnostic_data¶
The structured (interpolated) data for diagnostic cycles, as a dataframe. The format is similar to that of .structured_data. The datapath must be structured before this attribute is available.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file_with_diagnostic.071")
datapath.autostructure()
print(datapath.diagnostic_data)
# Out:
voltage test_time current ... step_type discharge_dQdV charge_dQdV
0 2.700000 NaN NaN ... 0 NaN NaN
1 2.703006 NaN NaN ... 0 NaN NaN
2 2.706012 NaN NaN ... 0 NaN NaN
3 2.709018 NaN NaN ... 0 NaN NaN
4 2.712024 NaN NaN ... 0 NaN NaN
... ... ... ... ... ... ...
44434 2.782701 1958305.375 1.612107 ... 0 0.0 0.006379
44435 2.783219 1958305.375 1.612090 ... 0 0.0 0.006379
44436 2.783736 1958305.375 1.612073 ... 0 0.0 0.006379
44437 2.784254 1958305.375 1.612056 ... 0 0.0 0.006379
44438 2.784771 1958305.375 1.612039 ... 0 0.0 0.006379
[44439 rows x 16 columns]
*Datapath.diagnostic_summary¶
A summary of the structured diagnostic cycle data, as a dataframe. The datapath must be structured before this attribute is available.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file_with_diagnostic.071")
datapath.autostructure()
print(datapath.diagnostic_summary)
# Out:
cycle_index discharge_capacity ... paused cycle_type
0 1 4.711819 ... 0 reset
1 2 4.807243 ... 0 hppc
2 3 4.648884 ... 0 rpt_0.2C
3 4 4.525516 ... 0 rpt_1C
4 5 4.482939 ... 0 rpt_2C
5 36 4.624467 ... 0 reset
6 37 4.722887 ... 0 hppc
7 38 4.584861 ... 0 rpt_0.2C
8 39 4.476485 ... 0 rpt_1C
9 40 4.426849 ... 0 rpt_2C
10 141 4.529535 ... 0 reset
11 142 4.621750 ... 0 hppc
12 143 4.486644 ... 0 rpt_0.2C
13 144 4.391235 ... 0 rpt_1C
14 145 4.336987 ... 0 rpt_2C
15 246 4.459362 ... 0 reset
16 247 4.459362 ... 0 hppc
*Datapath.get_cycle_life(n_cycles, threshold)¶
Calculate the cycle life for capacity loss below a certain threshold.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file.071")
datapath.autostructure()
print(datapath.get_cycle_life())
# Out:
231
*Datapath.cycles_to_capacities(cycle_min, cycle_max, cycle_interval)¶
Get the capacities for an array of cycles in an interval.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file.071")
datapath.autostructure()
print(datapath.cycles_to_capacities(cycle_min=50, cycle_max=200, cycle_interval=50))
# Out:
cycle_50 cycle_100 cycle_150
0 2.020498 1.981053 1.965753
*Datapath.capacities_to_cycles(thresh_max_cap, thresh_min_cap, interval_cap)¶
Get the number of cycles to reach an array of threshold capacities in an interval.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file.071")
datapath.autostructure()
print(datapath.capacities_to_cycles())
# Out:
capacity_0.98 capacity_0.95 capacity_0.92 capacity_0.89 capacity_0.86 capacity_0.83 capacity_0.8
0 76 185 231 231 231 231 231
*Datapath.is_structured¶
Tells whether the datapath has been structured or not.
from beep.structure import MaccorDatapath
datapath = MaccorDatapath.from_file("/path/to/my_maccor_file.071")
print(datapath.is_structured)
# Out:
False
datapath.structure()
print(datapath.is_structured)
# Out:
True
Making your own BEEPDatapath¶
If your cycler is not already supported by BEEP, you can write a class for structuring its data with BEEP by inheriting BEEPDatapath and implementing one method: from_file.
from beep.structure import BEEPDatapath
class MyCustomCyclerDatapath(BEEPDatapath):
"""An example of implementing a custom BEEPDatapath for your own cycler.
"""
@classmethod
def from_file(cls, filename):
# Load your file from the raw file filename
data = pd.read_csv(filename)
# Parse the raw data
# The raw data must adhere to BEEP standards. See the beep/conversion_schemas for the list of canonical data columns the raw data dataframe must posess.
# Your own code for converting the raw data to contain BEEP columns
data = convert_my_custom_cycler_data_to_BEEP_dataframe(data)
# Parse the metadata using your own code
# Metadata must return a dictionary
# Should preferably contain "barcode", "protocol", and "channel_id" keys at a minimum.
metadata_filename = filename + "_metadata"
metadata = my_metadata_parsing_function(metadata_filename)
# Store the paths in a dictionary
paths = {
"raw": filename,
"metadata": filename + "_Metadata"
}
return cls(data, metadata, paths)
Your custom datapath class can create new methods or override existing BEEPDatapath methods if needed.
Once you have written your custom class's from_file method, all the existing behavior of BEEPDatapath should be available, including
structure()validate()autostructure()pathsraw_datastructured_summarystructured_datadiagnostic_data- etc.
Electrochemical Impedance Spectra¶
More documentation for EIS coming soon!
Structuring compatibility with processed legacy BEEP files¶
Both legacy and *Datapath processed (structured) files saved as json should load with *Datapath.from_json_file, but the capabilities between files serialized with legacy and files serialized with newer BEEPDatapath files will differ.
The main discrepancy is that legacy files cannot be restructured once loaded. All of BEEPDatapath's other structured attributes and properties should function for legacy files identically to those serialized with newer BEEPDatapath.
See the auto_load_processed documentation for more info on loading legacy processed BEEPDatapaths.
Top-level functions for structuring¶
Aside from the CLI (shown in the command line interface guide, BEEP also contains lower-level python functions for helping loading and structuring many cycler output files from different cyclers.
auto_load¶
Auto load will look at the file signature of a raw cycler run output file and automatically load the correct datapath (provided the cycler is supported by BEEP).
from beep.structure import auto_load
arbin_datapath = auto_load("/path/to/my_arbin_file.csv")
print(arbin_datapath)
# Out:
<ArbinDatapath object>
maccor_datapath = auto_load("/path/to/my_maccor_file")
print(maccor_datapath)
# Out:
<MaccorDatapath object>
auto_load_processed¶
Automatically loads the correct datapath for any previously serialized processed (structured) BEEP file.
While processed run .json files serialized with *Datapath classes can be loaded with monty.serialization.loadfn, processed files serialized with older BEEP versions may not work with loadfn.
auto_load_processed will automatically load the correct datapath, even for legacy BEEP processed .json files, though the functionality of these datapaths is restricted. For example, legacy datapaths
cannot be restructured.
from beep.structure import auto_load_processed
arbin_datapath_processed = auto_load_processed("/path/to/my_processed_arbin_file.json")
print(arbin_datapath_processed)
# Out:
<ArbinDatapath object>
processed_datapath_legacy = auto_load_processed("/path/to/my_legacy_neware_file")
print(processed_datapath_legacy)
# Out:
<NewareDatapath object>